Dashboard for the Inference and Analysis of Networks from Expression data 
DIANE is a shiny application for the analysis of high throughput gene expression data (RNA-Seq). Its function is to extract important regulatory pathways involved in the response to environmental changes, or any perturbation inducing genomic modifications.
We designed this tool to process, explore, and perform advanced statistical analysis on multifactorial expression data using state of the art methods. It includes :
-
Raw count data pre-processing and sample-wise normalization
-
Customizable differential expression analysis
-
Gene ontology enrichment analysis for model organisms
-
Expression based clustering in the framework of Poisson Mixture Models, and characterization of those clusters with generalized linear models and GO enrichment analysis
-
Machine learning based Gene regulatory network inference
All of the features in DIANE are accessible via a single page shiny application that can be locally launched, or accessed online at https://diane.ipsim.inrae.fr{.uri}.
The steps should be performed in the order of the different tabs. For instance, before running clustering or network inference, differential expression should be performed first. The figure above summarizes DIANE’s main workflow.
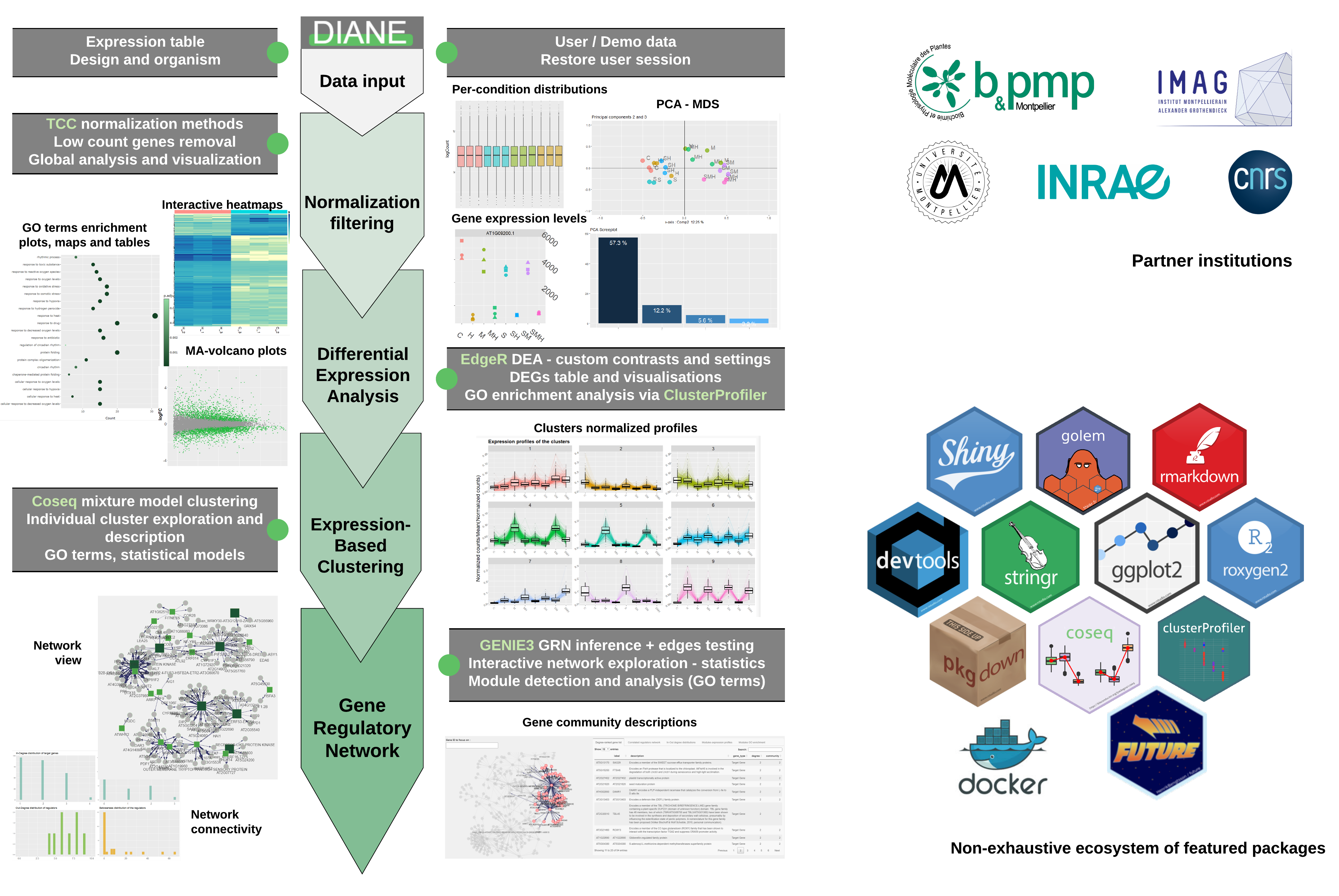
For more advanced users, all server-side functions in DIANE are exported so they can be called from R scripts.
Fore more information, please find full documentation and examples in the github page https://oceanecsn.github.io/DIANE.
Once the application is launched, if the resolution poorly fits your screen, you can adjust it with the keyboard shortcuts ctrl + or ctrl - (use cmd on Mac).
Please report any bug or suggestion via github or at oceane.cassan@cnrs.fr.
To cite DIANE in publications use:
Cassan, O., Lèbre, S. & Martin, A. Inferring and analyzing gene regulatory networks from multi-factorial expression data: a complete and interactive suite. BMC Genomics 22, 387 (2021). https://doi.org/10.1186/s12864-021-07659-2
A BibTeX entry for LaTeX users is
@Article{cassan2021Inferring,
title = {Inferring and analyzing gene regulatory networks from multi-factorial expression data: a complete and interactive suite},
author = {Océane Cassan and Sophie Lèbre and Antoine Martin},
journal = {BMC Genomics},
year = {2021},
volume = {22},
number = {387},
url = {https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-021-07659-2}}
Use DIANE locally
To use DIANE locally, download and install DIANE in your R console as follows (you need the remotes package installed) :
remotes::install_github("OceaneCsn/DIANE")
DIANE relies on R 4.0.0, available for all OS at https://cloud.r-project.org/.
You can then launch the application :
library(DIANE)
DIANE::run_app()
License
Copyright (C) 2020 Oceane Cassan
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.
Deploy DIANE on your server
We provide a solution based on Docker and Shiny server to deploy DIANE on any linux server, just as it is at https://diane.bpmp.inrae.fr. To do so, see the following command line instructions.
Get DIANE source code via Git :
git clone https://github.com/OceaneCsn/DIANE.git
Install Docker engine, as described in the Docker docs.
Go to DIANE’s folder.
First, you can change the default settings for the dockerized shiny-server by editing the file shiny-customized.config (like changing the port, the user to run with, and more)
Now let’s build the image, that we’ll name diane, from the Dockerfile (superuser rights required).
cd DIANE
docker build -t diane .
This might take a while. You can check that the container image was built with docker images. Then, you can start the container diane, by setting appropriately the following options in the above command:
/path/to/app/on/host/ is the path to DIANE on the host, that is to say the location where you cloned it, containing the app.R file. /path/to/logs/on/host/ is the folder you want to store your app logs.
-p 8086:8086 is the port to use, change the first 8086 to use another one on the host.
--user shiny allows to run as non root, with the shiny user that must have been created before, and granted rights to the folder /path/to/app/on/host/logs and /path/to/logs/on/host/.
-d --rm are options for the detached mode.
Plus, in the following example, a session of DIANE will be allowed to use 16 CPU cores :
docker run -d --cpus 16 --user shiny --rm -p 8086:8086 -v /path/to/app/on/host/:/srv/shiny-server/ -v /path/to/logs/on/host/:/var/log/shiny-server/ diane
You can check that the container is running with docker ps.
Authors : Océane Cassan, Antoine Martin, Sophie Lèbre
Dev : Océane Cassan, PhD Student at IPSIM (Institute for Plant Sciences in Montpellier) research unit, SUPAGRO Montpellier, with contributions from Alexandre Soriano.
Upload expression data and experimental design
Expression file upload
Preview of the expression matrix
Design and gene information files
Data filtering and normalisation
Settings
Normalization
Low counts filtering
Minimal gene count sum accross conditions :
Explore normalized gene expression
Differential expression analysis
Settings
Estimation of disperion
Conditions to compare for differential analysis :
Expression based clustering
Settings
Mixture models clustering
Analyse the genes of a specific cluster
Expression profiles
Network inference
Inference Settings
GENIE3 Gene regulatory network inference
Or choose your own :
Thresholding settings
Without thresholding, we would obtain a fully connected weighted graph from GENIE3, with far too many links to be interpretable. In order build a meaningfull network, this weighted adjacency matrix betwen regulators and targets has to be sparsified, and we have to determine the N higher regulatory weights that we consider significant.
As biological networks are known for their pronounced sparsity, considering all possible regulator-target pairs would be of very little interest. Studies found that the typical values of density (ratio between the number of edges, and the number of edges of the complete graph) in state of the art biological networks lie approximately between 0.1 and 0.001. We therefore propose to create a first graph, topologically consistent with biological network standards, which will be further refined by statistical testing. During the testing phase, the null ditributions of regulatory weights are estimated by the permutations, thus providing an empricial p-value for the observed importance.
Network analysis
Click on any node (gene) to get its description and network information
Legal mentions
For DIANE’s online version <https://diane.bpmp.inrae.fr>{.uri}
Website publication
Webmaster and content publisher : Océane CASSAN
Website hosted at : Campus INRAE / SupAgro – IBIP – UMR BPMP
Bât. 7 – 2 place Pierre Viala
34060 Montpellier cedex 2
Affiliations
DIANE was developped with the following academic organisations support:
INRAE
Établissement public à caractère scientifique et technologique
147, rue de l’université
75338 Paris cedex
07 01 42 75 90 00
CNRS
3, rue Michel-Ange
75794 Paris cedex 16 - France
+33 1 44 96 40 00
BPMP and Agro Institute
Campus INRAE / SupAgro – IBIP – UMR BPMP
Bât. 7 – 2 place Pierre Viala
34060 Montpellier cedex 2
University of Montpellier
163 rue Auguste Broussonnet
34090 Montpellier
Data privacy and cookies
All data submitted to DIANE for analysis will remain confidential as it is not stored on the server side.
DIANE doesn’t collect any private information on users. It only makes use of technical cookies as well as anonymous usage data.
License
Copyright (C) 2020 Oceane Cassan
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.
Version changes in DIANE
1.0.1 : DIANE is released
Version at the moment of the official BMC publication (May 26, 2021).
1.0.2 : Minor bug fixes and improvements.
Improvements :
- Citation was added
- Gene IDs can now have blank space round them in the exploration of specific genes profiles
- MDS was removed
- PCA can now handle expression data with 4 or less experimental conditions
- This document was added to the interface in a new tab
Bug fixes :
- Custom annotation/gene information file now allows duplicated genes
- Custom annotation/gene information now can only have columns named label and/or description
- DEA csv download does not require annotations any more
1.0.3 : Minor bug fixes and improvements.
- Fixed error in DAINE’s programming interface (wrong value of the defaut “conditions”” argument in normalize function) 22/11/2021
- Used namesapce operator in data import module to speed up loading
1.0.4 : Minor changes, mainly aesthetic, with no impact one DIANE’s main functions and backend. Changes are from merging pull request #36 from Alexandre-So.
-
Some code based on bootstrap grid system has been rewrite. This avoid the grey (anthracite) background to show up at the bottom of some pages, and improve shinydashboardPlus::box alignment between different dashboard tabs. This also increases a little bit the width of most of the shinydashboardPlus::box.
-
Numbers in the DEA table show less digits when displayed in DIANE (up to 10-3). The downloaded table is not affected.
-
When a row is clicked in the DEA table, display a popup with RNAseq count associated with the related gene. (Very cool feature, worth the highlight)
-
The little loading square position is now based on user screen resolution. The position can still be improved. Used CSS code is in app_ui.R.
-
The little help buttons in the import dataset page have been moved to new locations. This can be done for other tabs.
-
Add “y axis” scrollbar to some tables which were exceeded screen boundaries in low resolutions.
-
Update contact email to new cnrs one.
1.0.5 : Minor changes and update caused by breaking changes in dependency
-
rfPerumte was updated and code had to be change to comply with new function names
-
The vignette was improved (mention of seed, and rendering adjustements)
1.0.6 Fixes
-
Changed url to diane.ipsim.inrae.fr added license to welcome pages
-
Fixed cutom gene info file upload in the case of splicing aware gene IDs in expression file
-
Fixed not displaying go results for custom org in network communities
-
In draw expression levels, genes filtered because of low expression can still be viewed
January 8, 2024 - Fix
Fix a bug with normalization : Normalization was using total count instead of the normalization factors computed by the chosen method.